Fundamentals of Bioinformatics
About Fundamentals of Bioinformatics
Fundamentals of Bioinformatics (02-604) is a 12-unit course for graduate students at Carnegie Mellon University, and it is a requirement for students in the MS in Automated Science program.
How do we find potentially harmful mutations in your genome? How can we reconstruct the evolutionary Tree of Life? How do we compare related genes from different species? These are just three of the major questions in modern biology that can only be answered using computational approaches.
This delves into computational ideas used in biology as well as let students apply existing resources that are used in practice every day by bioinformatics professionals. The course also offers an opportunity for students who possess an introductory programming background to become more experienced coders in a biological setting. And it requires all students to complete an open-ended project on a topic of their choice related to bioinformatics.
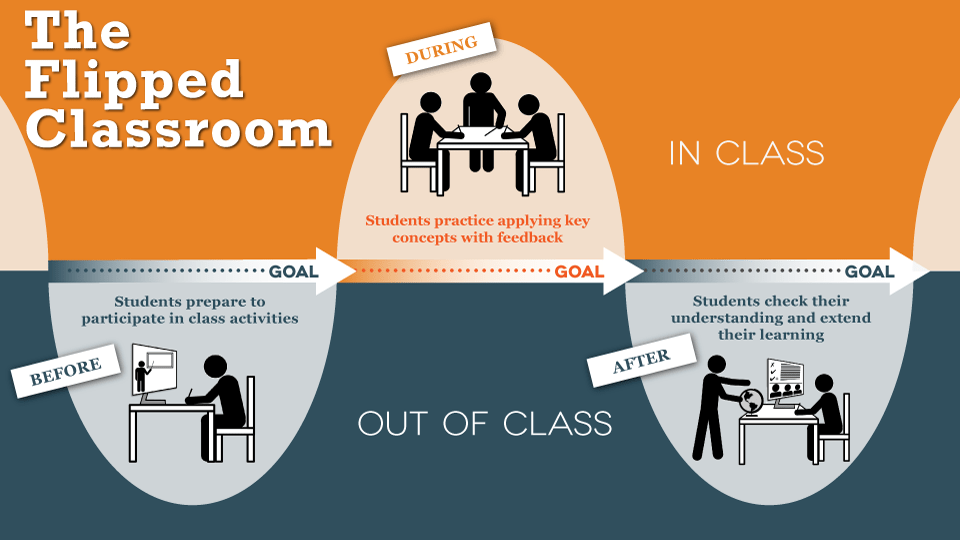
Fundamentals of Bioinformatics is built on a flipped model of teaching, which I describe in the “Course Philosophy” section below.
Student Testimonials
“Phillip is a great instructor who obviously cares about the success of his students. After taking a few of his classes, I’ve found that he sometimes has a nontraditional pedagogical approach, but everything is well thought out and done for good reason. This course featured a “flipped classroom” approach, where most of the material was encountered outside the classroom in an interactive online textbook, while class meetings involved mostly Q&A, small-group challenge questions, and discussion. I found the online content to be very helpful for understanding the material.”
“Love the course.”
“Dr. Compeau demonstrated a great passion and expertise in the subject he taught. Also he has fantastic teaching styles and techniques which is a not a common quality for university professors if not rare. The overall materials are intriguing to learn, have a good converge on many topics in bioinformatics as well as into the necessary depths of all the algorithms to implement. I learned a lot from this course, both from theory, and hands-on experience with the state-of-art bioinformatics software. I truly recommend it to others as by far this is my favorite course at CMU. The interactive textbook is a also a gem, designed in a very informative yet not boring way.”
“There are many teachers, instructors and professors out there, and then there are the real EDUCATORS. Phillip is a genuine educator.”
“The most beneficial course I’ve ever taken.”
Prerequisites
There are no formal prerequisites, but the course assumes an introductory background in biology, some algorithmic thinking, and a strong foundation in programming, such as that provided by Programming for Scientists.
Course Philosophy
Fundamentals of Bioinformatics was a finalist for a Teaching Innovation Award at Carnegie Mellon, and it was a subject of an invited education article I wrote based on a blog post describing my experience teaching the course.
The central motivation for how this course operates is based on the “2 Sigma Problem”, a paradigm of educational psychology introduced by Benjamin Bloom in 1984. This remarkable principle, which has been verified by student performance, states that students achieve a two standard deviation improvement in learning performance when they are tutored individually compared to a standard lecture in a 30-student classroom — let alone a 100-student classroom!) The reasoning behind the 2 Sigma Problem is that 30 students learn different material in different ways and at different paces, but the traditional lecture is completely blind to this conundrum. In Bloom’s words,
Teachers are frequently unaware of the fact that they are providing more favorable conditions of learning for some students than they are for other students.
How, then, should a course be run? The instructor of any 30-student class unfortunately does not have time to tutor every student in the course individually. Instead, we must strive for extreme class discussion and “mastery learning”, in which a subject is understood perfectly before the next topic is covered.
These principles are not enacted by most courses because most courses do not have the time or resources to enact them. Fundamentals of Bioinformatics differs because it is powered by an interactive version of the popular Bioinformatics Algorithms project that I co-developed with Pavel Pevzner.
The course therefore operates on a “flipped” model, in which students read and complete assessments in the interactive text (as well as watch optional lectures) outside of normal class time. This allows for class time to be used not for transmissive lectures but rather for in-depth guided discussions with students working together in small groups.
Fundamentals of Bioinformatics Course Details
Course Meetings
Fundamentals of Bioinformatics meets once per week for approximately 150 minutes. Each weekly meeting consists of some or all of the following items:
- Resolving common questions from the week’s reading.
- A mini-lecture on a topic of interest connected to the week’s topic.
- Discussion questions probing understanding on the week’s topic.
- Challenge questions that extend the knowledge obtained of the topic to a new area.
- Questions encouraging students to start thinking about the next week’s topic.
The final three items are typically completed in randomly assigned groups of 4-5 students with instructor and TA support.
Interactive Text
The course is powered by an interactive version of Bioinformatics Algorithms. Every week, students are assigned a reading from this book (online lectures from our YouTube channel are provided as well).
Programming assignments are integrated directly into the interactive text and are due at the end of every week (typically the week after a class discussion). These assignments are language neutral, so that students can complete them in any programming language they choose.
Quizzes
To ensure understanding, a weekly comprehension quiz is provided along with each week’s reading, due the night before class.
Weekly Write-ups
On the night before class, each student submits a write-up detailing their experience with that week’s reading.
The write-up includes the following components directed at the students.
- A report of every difficulty that you had in understanding the material, documenting the precise location where the misunderstanding occurred. How did you resolve the misunderstanding?
- A precise formulation of every remaining question that you have regarding understanding the material. A poorly formulated question would be, “I don’t understand how Algorithm X works; can you please explain it again?” A better formulated question would isolate the source of the misunderstanding as closely as possible, indicating the specific location in the text that caused the misunderstanding (e.g., “If I input the data in Figure Y into Algorithm X, I notice that in line Z, I obtain a different result to the output in Figure Y”). Questions should not be easily answered in the text.
- Any remaining questions that you have about the material that you’re curious about.
Software Challenges
Software challenges give students the opportunity to learn the basics of commonly used bioinformatics resources (such as BLAST, MEGA, MEME, SPAdes, etc.) These challenges reinforce the algorithms by showing how they are applied to real data in the software used every day by biologists around the world.
Fundamentals of Bioinformatics Course Projects
The core material in Fundamentals of Bioinformatics focuses on the algorithmic side of computational biology: modeling biological problems computationally, formulating algorithms to solve our problems, and then reflecting on these algorithms. To balance this theory with some practice, we give students the chance to explore a practical challenge of their own choosing by running a computational analysis on a real biological dataset and then interpreting the results. Students complete this project individually or in a small team of their own choosing, and they receive frequent feedback on their work throughout the semester.
There are two main directions for the project. First, students may implement an algorithm solving a biological problem and then use this algorithm to analyze a biological dataset. Second, students may analyze the dataset only using existing software. In either case, the primary expectation for the project will be based on the analysis performed.
For example, if students were to write your own genome assembler for real biological data (a very challenging task to undertake), then it might suffice to apply code to a bacterial read dataset and compare the resulting assembly and its statistics to those of an existing program. But if only using existing software to assemble a genome, then we would expect students to take the project into additional directions to have a complete analysis.
Fundamentals of Bioinformatics Project Ring of Honor
The following projects are an example of exemplary student work.
Expression pattern detection and classification of two types of lung cancer, LUAD and LUSC
Project author: Xin Wang
group4xin_78574_8093121_02604-PaperLong-Read Genome Assembly: A Case in Study in Python
Project authors: Dylan Estep, Ethan Gaskin, Akshat Gupta, and Zhen Yang
groupdsmrtypants_96001_11176599_project-report-group-D-1Bacterial DNA in the Spotlight
Project author: Manjot Nagyal
groupfjoisalljoneedsstrainsformetagenomicanalysis_LATE_111003_11178119_02-604_Bioinformatics_Project_FinalReport