PreCollege Program in Computational Biology: A Unique Voyage

About the PreCollege Program
The PreCollege Program in Computational Biology is part of Carnegie Mellon’s suite of PreCollege programs, a collection of excellent summer educational programs designed to give high school students exposure to unique educational experiences on campus.
Despite the computational revolution within modern biology, most students who take biology in high school obtain no appreciation for the fact that biology has become a data science. To fill this unmet need, Josh Kangas and I founded our program to show students the interplay between experimentation and computation in life sciences.
Our area of focus is on microbe genomics and metagenomics. Specifically, we build code to analyze DNA taken from both publicly available SARS-CoV-2 datasets as well as from samples of river water that we captured (along with our students) from Pittsburgh’s three rivers.
Josh and I have spent a great deal of time and effort in the successful administration of this program, and we care deeply about recruiting as many underrepresented students as possible. I say more about our efforts in my Administration statement.
Student testimonials
“I’m so grateful to Mr.Compeau and Mr.Kangas for everything they did for me this summer. I learned SO much thanks to their instruction and they never hesistated to help me when I was confused. I had lots of questions and they never made me feel like that was a bad thing. I cannot thank them enough.”
“The Pre-College Computational Biology course was extremely fulfilling, fun, and eyeopening. I came in with high expectations from the program and it is safe to say that I learned and enjoyed the course much more than I thought I would. The practical experience and hands on exercises made the program much more interactive and demanding, but led to a much better understanding of the algorithms and methods we learned. Not only did the program fuel my interest in Computational Biology, much of the material covered is now directly applicable. During the program, I obtained a research position at a local university and am now using many of the algorithms that I learned during the Comp. Bio program everyday. Professors Kangas and Compeau were really helpful in teaching Computational Biology in all aspects, from the programming, to the implementation to the data analysis and visualization. By using real world data instead of artificial data (to an extent), the program was a lot more fulfilling as it felt like we were really practicing real Comp. Bio techniques.”
“Mr. Compeau always respects and considers a student’s difficulty. He would asked privately through the zoom chat box to see if students are comfortable answering questions in class instead of just cold-calling.”
“Phillip is the BEST teacher I have had in my life. Within my first week of pre-college, I learned as much as I had in a semester of my AP Computer Science course in high school. Not only did I learn the material, but I processed and retained it, and was always curious to ask more. The way [Phillip] connects seemingly unconventional real-world problems to computational biology is incredible because it helps us, as students, gain a very good grasp on conceptual knowledge. For each problem presented to us, Phillip always challenged us and encouraged us to speak up and think, even if we were wrong. A great teacher, and deserves all of the praise I can give him.”
“Dr. Compeau was the best STEM educator I have met. Not only did he thoroughly take us through more than years worth of CS content in a staggering three days, but he really allowed the building of these core fundamentals to develop our logical/programming knowledge, as he would introduce lines of logic while we were programming. He also taught us very important programming skills, such as subroutines, recursion, dynamic programming, etc. I truly had an amazing experience in the CS lab.”
Prerequisites
We are looking for students who are both strong in mathematics and who are interested in learning about an aspect of biology that they have probably never contemplated, and that truly is at the cutting edge of biological research. Students can demonstrate both of these prerequisites in a number of ways.
We also know that, despite many excellent outreach efforts, computer science is still not offered as a course of study for most high school students in America. For this reason, it was a founding principle of our program that we not require our students to have any programming experience, and we provide required preparatory materials to all admitted students taken from my Programming for Lovers project.
PreCollege Program Curriculum
Under normal circumstances, students spend four hours per day with me in a computer lab, coding in small groups with instructor guidance to solve computational challenges in a hackathon model, and then applying the code that they write to our datasets. In this way, the program implements active learning, so that students are doing as much as possible to facilitate their learning (I say more about active learning as a thread running through all of my teaching in my statement of teaching philosophy).
Students then spend four hours per day in the lab with Josh, conducting experiments related to analyzing bacterial samples taken from river water. From 2022 onward, we will be leveraging CMU’s unparalleled automated laboratory infrastructure to allow students to drive instruments robotically and at much greater scale.
In 2020 and 2021, we ran an online version of our precollege program that allowed the program to double in size and that focused exclusively on the computational side of the program.
Below, I provide an outline of the curriculum for my half of the course, in which students learn about fundamental algorithms for biological data analysis and code implementations of these algorithms in groups.
Module 1: Quantifying Metagenomic Diversity
Once we have a metagenomic sample (e.g., many fragments of DNA isolated from a bottle of river water), we ask two questions:
- How diverse is this sample?
- How does this sample compare to other samples?
The first notion of diversity (intra-sample diversity) is called the sample’s alpha diversity, and the second notion of diversity (inter-sample diversity) is called beta diversity.
Quantifying both alpha and beta diversity predate the era of cheap DNA sequencing, and there are quite a few functions, such as Simpson’s index and Bray-Curtis distance, that are easily accessible to high school students and that can be coded by beginners quite quickly and applied to real datasets to yield biological conclusions.
For example, we see that the alpha diversity of samples varies greatly by season, as shown in the following figure.
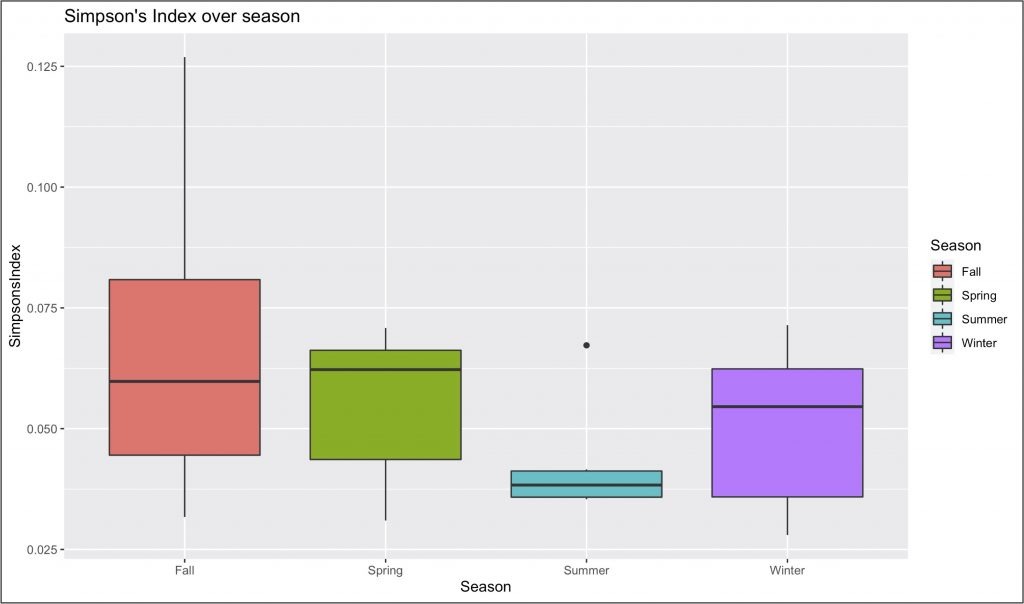
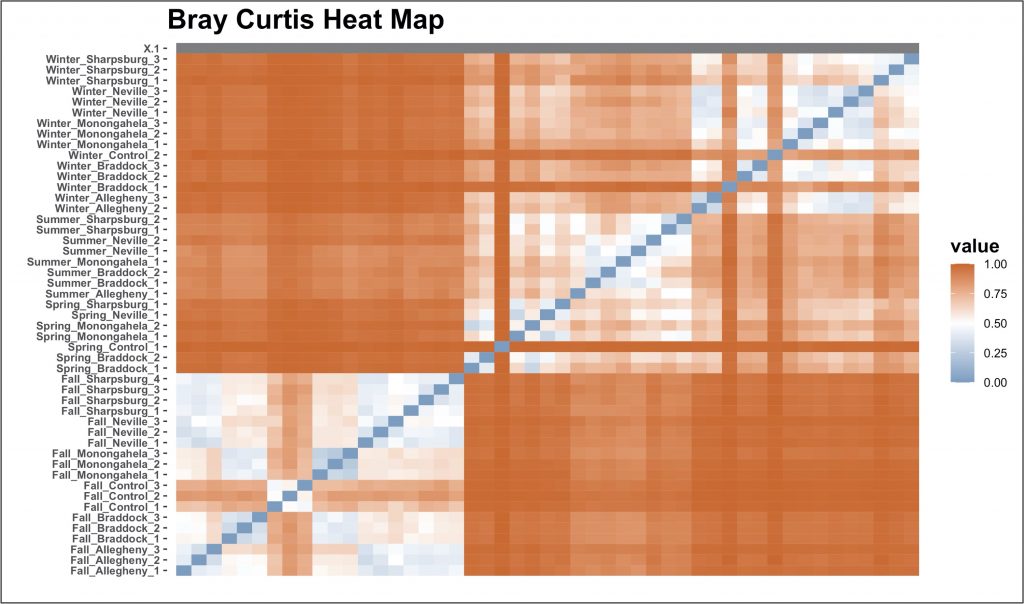
We also see that the beta diversity between samples heavily depends on season, as shown by the following “heat map” of locations.
Module 2: Comparing Biological Sequences
Once we have sampled fragments of DNA from a metagenomic sample, the next question we might ask is, “What organisms did they come from?” In other words, how can we compare given fragments of DNA against a database of organisms with known genomes?
A simpler question is how to compare two DNA strings (made up of the symbols “A”, “C”, “G”, and “T”). Once we have an algorithm for comparing two strings, we can generalize it into an algorithm for comparing a given DNA string against an entire database, and then apply our algorithm to figure out what is lurking in our rivers after all, as shown in the following bar charts.
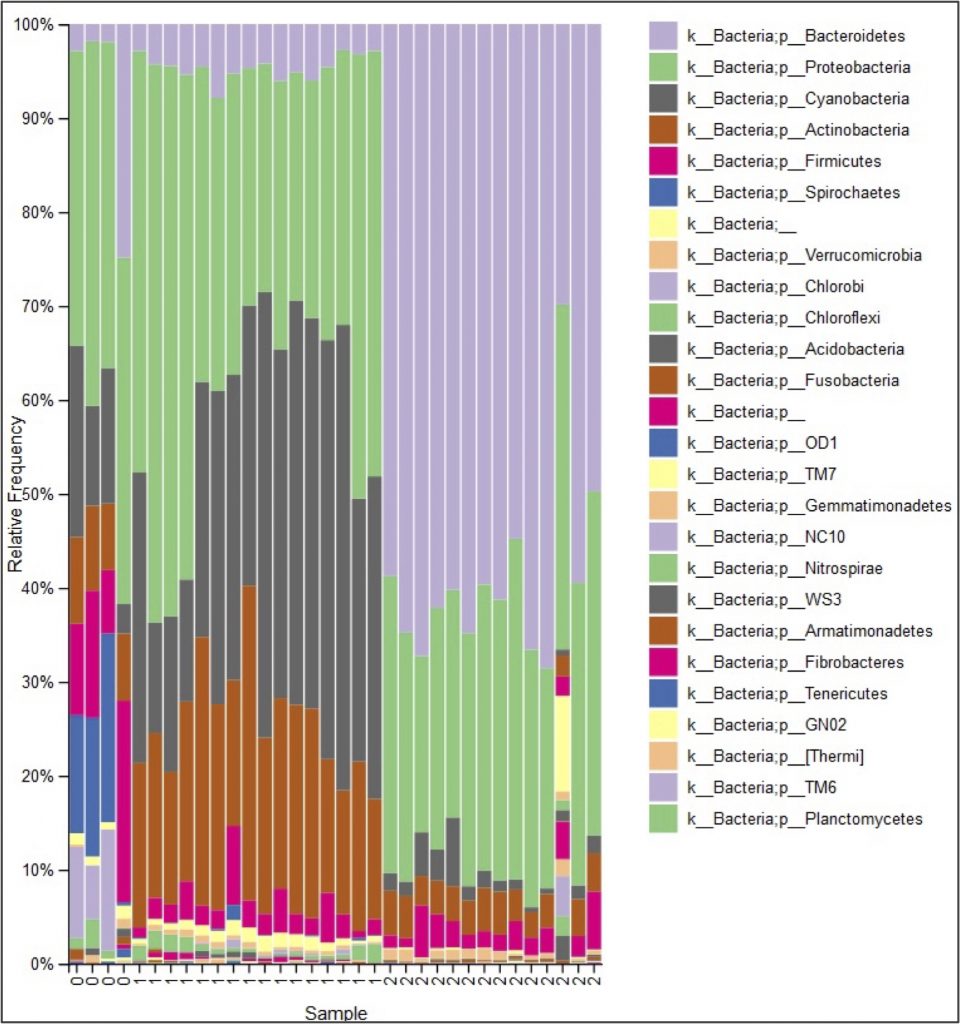
Module 3: Assembling Genomes
Another example of using fragments of DNA is in the context of genome sequencing, in which we read fragments of DNA sampled from many copies of the same genome (e.g., multiple copies of the SARS-CoV-2 virus, or multiple bacterial cells from the same species). Our goal is to use overlapping fragments of DNA to infer the original genome, as highlighted in the figure below.
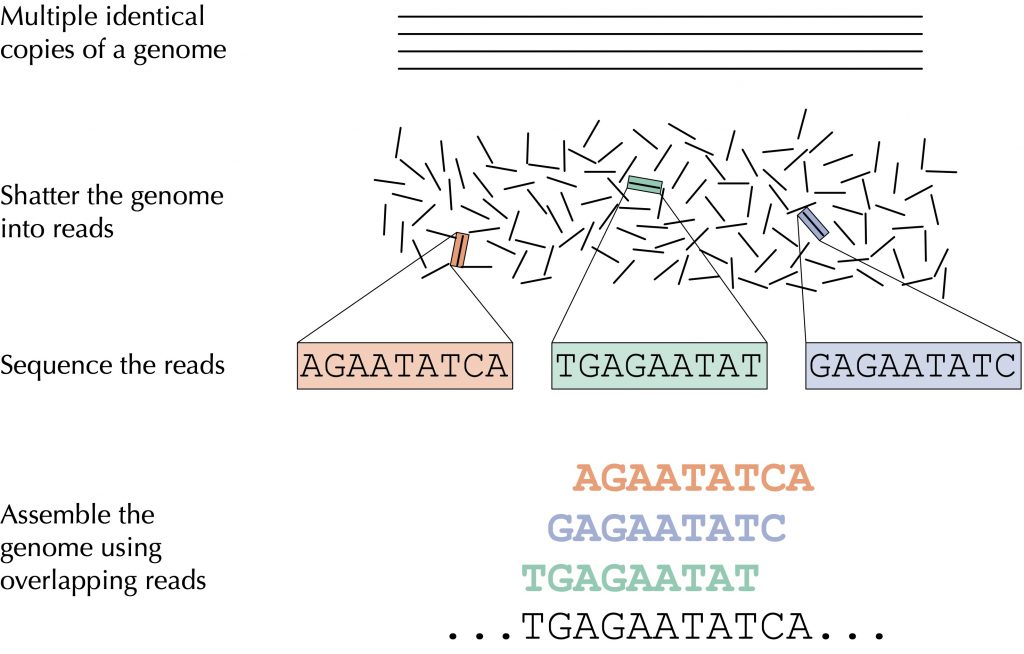
Building an efficient algorithm to overlap all these fragments into a contiguous genome is a very complex task, but it is one at the heart of computational biology. We then apply our algorithm to fragments of DNA captured from SARS-CoV-2. Can we place ourselves in the shoes of researchers at the start of the pandemic and sequence this viral genome?
Module 4: Finding Genes
Once we have sequenced a full genome, we possess a very rich dataset that gives a complete blueprint of an organism’s genetic identity. But what can we do with it?
The first thing that we might do is look for the genes lurking within an organism’s genome so that we can start to learn what purpose these genes serve and how the organism compares to others.
We build a simple but powerful gene finder, and we then see how our approach can be used to annotate the SARS-CoV-2 genome as shown in the figure below.
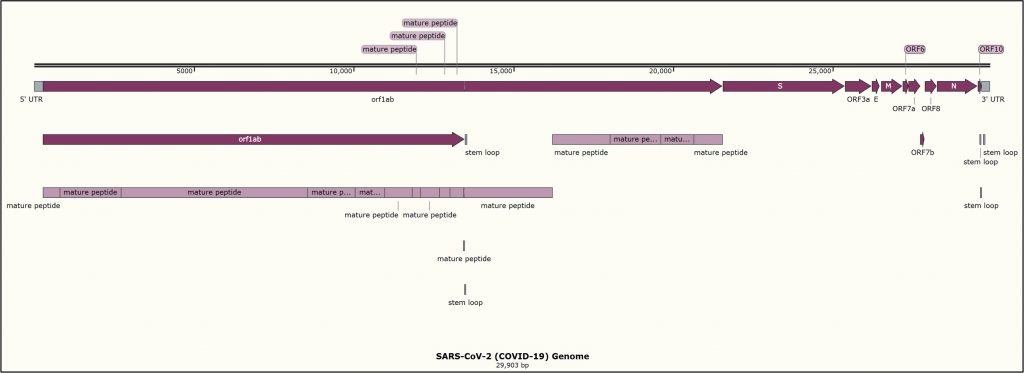
Module 5: Constructing Evolutionary Trees
The evolutionary tree is probably the most fundamental data structure in biology, dating back to a doodle in Darwin’s notebook. We discuss how one can design an algorithm that takes genetic data as input (e.g., the same gene taken from many different organisms) and outputs an evolutionary tree.
We then apply our algorithm to our metagenomic bacterial data to examine the taxonomy of bacteria in our samples, as well as build evolutionary trees for SARS-CoV-2 genomes sampled from patients in the same time frame, such as that one shown in the figure below for individuals from China, Italy, and the United States.
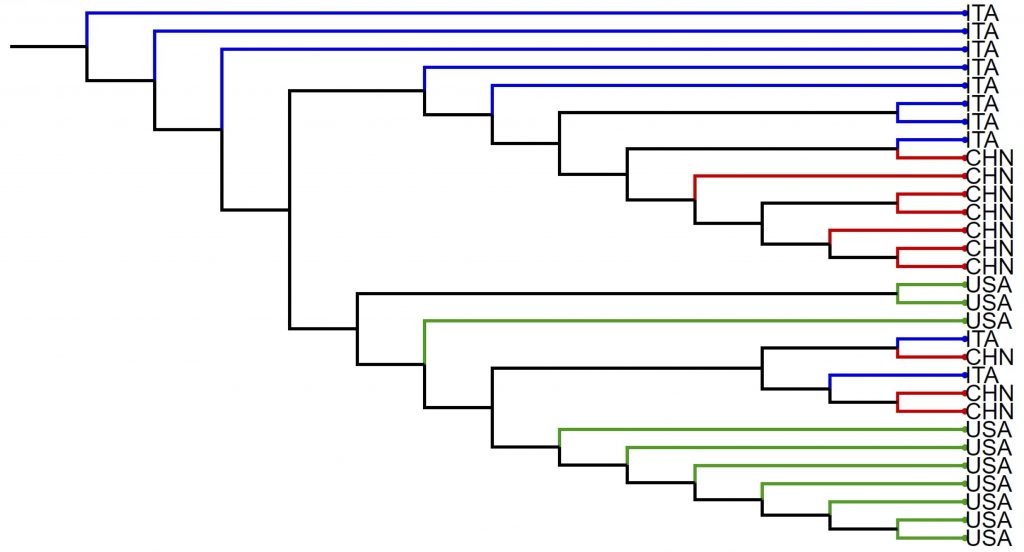
We then see that an evolutionary tree of coronaviruses allows researchers to identify viral variants in real time, as they arise. Once we can identify coronavirus variants, we turn our work to tracking these variants and seeing the seismic changes that happen within the human population in terms of percentages of variants over time.
