Essential Mathematics and Statistics
Motivation for Essential Mathematics and Statistics
In teaching master’s students within the computational biology department, I noticed that although our students were quantitatively talented, they lacked consistent foundation in the mathematical approaches needed for rigorous future coursework in machine learning and computational biology. Accordingly, I designed an accelerated foundational mathematics course called “Essential Mathematics and Statistics” intended for first-semester students in the MS in Computational Biology as well as the MS in Automated Science. (These students take the course concurrently with Programming for Scientists.) I co-taught the course with Seyoung Kim in its first offering in fall 2019.
The course is divided into two halves, with the first focusing on a variety of topics in mathematics and logic, and the second half focusing on statistics. I have taught the first half of this course, and I provide its curriculum below.
Course Description
Essential Mathematics and Statistics rigorously introduces fundamental topics in mathematics and statistics to first-year master’s students at Carnegie Mellon University as preparation for more advanced computational coursework. Topics are sampled from information theory, graph theory, proof techniques, phylogenetics, combinatorics, set theory, linear algebra, neural networks, probability distributions and densities, multivariate probability distributions, maximum likelihood estimation, statistical inference, hypothesis testing, Bayesian inference, and stochastic processes.
Students completing this course obtain a broad skillset of mathematical techniques and statistical inference as well as a deep understanding of mathematical proof techniques. They will have the quantitative foundation to immediately step into an introductory master’s level machine learning or automation course. This background will also serve students well in advanced courses that apply concepts in machine learning to scientific datasets, such as Computational Genomics or Automation of Scientific Research.
Student Testimonials for Essential Mathematics and Statistics
“We held our heads up high with you as the instructor and not coming from conventional ‘math’ background did not seem as scary anymore.”
“The part instructed by Phillip provided interesting and clear explanations of mathematical theories, which was very helpful.”
“Considering the amount of material that had to be covered and conveyed to the students in a short amount of time, I thought it was a great job.”
“I didn’t think I would enjoy math class, but I did enjoy yours! Thank you!”
Prerequisites
There are no formal prerequisites. However, we expect that students will have a strong foundation in high school mathematics and possess strong quantitative reasoning skills, as the course is taught at an accelerated pace.
Essential Mathematics and Statistics Curriculum
A curriculum of the mathematics half of the course is found below. Each module focuses on a single mathematical area, and the module is motivated by an application that is pulled from some area of biology. This feature makes the course relatively unique, as I am unaware of an introductory mathematical proofs course that focuses on scientific applications.
Module 1: Assembling Genomes (Graph Theory and Constructive Proofs)
Researchers assemble genomes from millions of submicroscopic overlapping fragments of DNA. We will see that graph theory, or the mathematical study of networks, is vital to solving this problem on real data.
Topics covered:
- Directed and undirected graphs
- Hamiltonian and Eulerian paths
- The Bridges of Königsberg
- Euler’s constructive proof for finding an Eulerian cycle
- Assembling genomes with de Bruijn graphs
- Pigeonhole principle
- Handshake lemma
Module 2: Two Gems from Ancient Greece (Proof by Contradiction)
Our focus is on two theorems whose proofs were discovered in ancient Greece well over two thousand years ago.
- √2 is irrational.
- There are infinitely many prime numbers.
In A Mathematician’s Apology, G. H. Hardy made the case for why these two theorems exemplify the elegance of mathematics. In Hardy’s Words:
They are “simple” theorems, simple both in idea and in execution, but there is no doubt at all about their being theorems of the highest class. Each is as fresh and significant as when it has discovered — two thousand years have not written a wrinkle on either of them. Finally, both the statements and the proofs can be mastered in an hour by any intelligent reader, however slender [their] mathematical equipment.
This module focuses on these two theorems, and how proof by contradiction is useful to both. Although every other module has a scientific focus, this module is about showing how beautiful mathematics for its own sake can be.
Topics covered:
- Integers, rational, and irrational numbers
- Relatively prime, prime, and composite integers
- Proof by contradiction
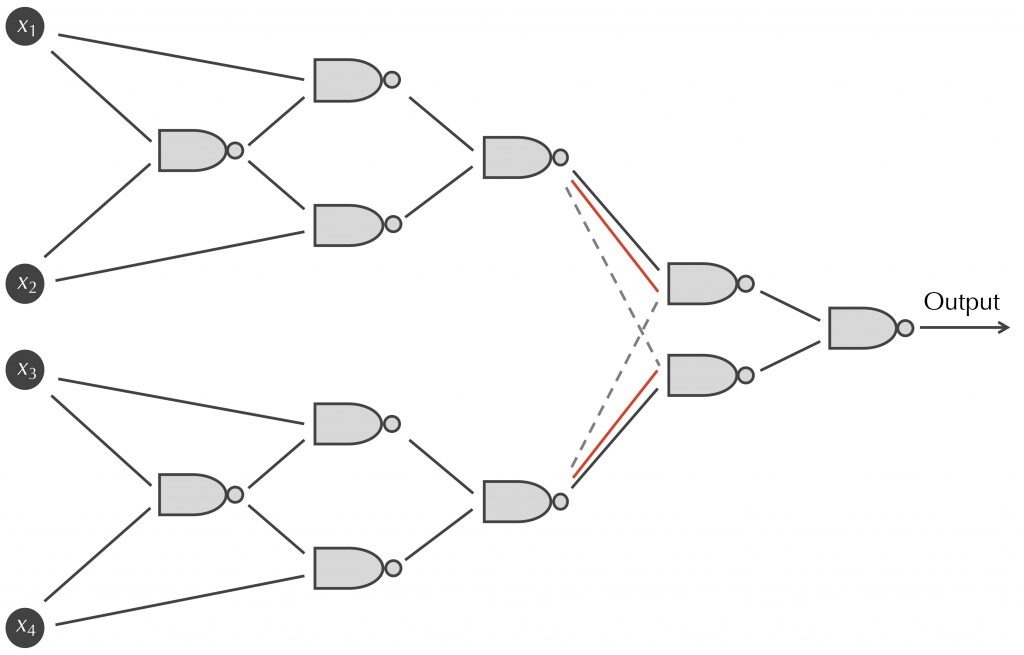
Module 3: The Evolution of Modularity and the Universality of Binary Neural Networks (Classical Logic)
Organisms form communities. Your body is divided into appendages, and your limbs have fingers and toes. Your organs each perform separate functions. Your tissues are all divided into cells. The cells themselves have specialized organelles and a nucleus. The nucleus contains DNA, which is divided into genes. Even genes divide into exons and introns, and the proteins that they encode comprise separate subunits called “domains”.
All of these different natural phenomena exemplify a concept called modularity, in which a system can be divided into components. An important attribute of modularity is that each component has many more interactions internally than it does with other components. The examples given in the previous paragraph indicate that modularity can be hierarchical as well.
Furthermore, modularity arises in the study of biological networks. To take one example, if we form a network whose nodes are the collection of proteins in a cell, and whose edges represent certain interactions between proteins, then we will observe many recurring subnetworks called “motifs”. To take another, networks of neurons in the body also tend to cluster into modules.
To study evolution, we could build a model in which objects are allowed to randomly mutate over a series of generations, with the most “fit” individuals retained in each generation to propagate. And yet until the 21st Century, no one was able to build such a model in which the most fit individuals at the end of the simulation exhibited modularity. How, then, has modularity evolved as such an important feature across the natural world?
We introduce neural networks and show the result of the beautiful Kashtan-Alon experiment, in which neural networks were allowed to evolve in an attempt to achieve a changing “fitness function”. To do so, we need to understand the foundations of logic and neural networks underlying this result.
Topics covered:
- McCullough-Pitts neurons
- Truth tables of logical connectives
- Perceptrons
- Linear separability and XOR
- DeMorgan’s Laws
- Logical equivalence
- Proof by cases
- Neural networks with hidden layers
- The universality theorem for networks of perceptrons and binary functions
- The ability of NAND gates to represent all binary functions
- The Kashtan-Alon algorithm
Module 4: Trees (Induction and Combinatorics)
The evolutionary tree is a fundamental construct in biological data analysis and is used in various applications, from understanding the spread of SARS-CoV-2 during a pandemic to building a “tree of life” categorizing life on Earth.
Researchers have developed many algorithms for evolutionary tree construction, but we will discuss the perfect phylogeny algorithm and apply it to the context of genotyping many humans with a collection of different genetic markers.
The explanation of why the perfect phylogeny algorithm works is powered by a mathematical principle of induction.
We also ask, “How many tree structures are there?” This question leads us to a beautiful result called Cayley’s theorem.
Topics covered:
- Trees, rooted and unrooted
- Product rule for combinatorics
- Combinations and permutations
- The binomial theorem
- Triangular and pyramid numbers
- Theorems about trees
- Bijective combinatorics
- Cayley’s formula
Module 5: More Powerful Neural Networks for Deep Learning (Real Analysis)
In a previous module, we introduced neural networks in which the inputs and outputs at each node were constrained to be either 0 or 1. These networks become much more powerful when we allow neurons’ inputs and outputs to be arbitrary real numbers. But what are the applications that we would hope to turn them to?
A seminal problem in data science is taking a collection of objects as input and attempting to “classify” them as one of a collection of n different categories as output. We can apply neural networks to this problem by converting each object to a vector of real numbers. We then can build a network of artificial networks with n output neurons, and change the “weights” assigned to each neuron in the network so that only the output neuron corresponding to a given object’s class fires when this object is used as input.
This simple approach powers much of what is called deep learning. But why does it work? Why would a simple network of artificial neurons with no intelligence be able to classify complicated objects (say, biological images) into categories and beat a human?
Funnily enough, the answer to this question in large part depends on some critical mathematics from the field of real analysis, the study of the real numbers making up the number line.
Topics covered:
- The problem of classification
- Training and test data
- Limits
- Continuity
- Arithmetical combinations of continuous functions
- Intermediate Value Theorem
- Extreme Value Theorem
- Approximability of all continuous functions by neural networks
Module 6: Genotyping (Linear Algebra)
In a previous module, we discussed the problem of genotyping individuals to reconstruct an evolutionary tree. In practice, however, individuals demonstrate a great deal of admixture of human populations. The question is how to deconvolve this admixture and provide an individual with a clear picture of how their individual ethnicity derives from these populations.
To do so, we will see that we need linear algebra, the branch of mathematics concerned with the study of matrices and systems of linear equations.
Topics covered:
- Curse of dimensionality
- Vectors and arithmetic operations
- Span and linear combinations
- Linear dependence and independence
- Bases
- Linear transformations
- Matrix arithmetic
- Inverse matrices and invertibility
- The Invertible Matrix Theorem
- Change of basis
- Diagonalizability and orthogonal diagonalizability
- Singular value decomposition (principal components’ analysis)
